Links to applications using or interfacing with Packmol
Cellulose BuilderCreating crystalline materials is not a Packmol task and can be tricky. This package builds cellulose fibers in crystalline forms with user determined sizes and geometries, which can then be solvated with Packmol. [Home-Page]
Amber Tools
Packmol has been incorporated into Amber Tools, with special scripts to build lipid bilayers! [Home-Page]
Molecular Dynamics Studio
This is a collection of software modifications created to integrate NanoEngineer-1, PACKMOL and MSI2LMP for the purpose of easily creating molecular dynamics cells. NanoEngineer-1 is a molecular CAD software written by Nanorex and provides the user an easy way to create molecules, while the software modifications allow the user to type atoms using multiple force fields. PACKMOL can generate a random collection of molecules using the molecule templates from NanoEngineer-1 thus providing the initial MD cell. Modifications to PACKMOL allow the atom type data to be passed through to the MSI2LMP software. MSI2LMP creates a LAMMPS input file based on class I or class II force fields. [Home-Page]
Moltemplate (http://www.moltemplate.org)
Moltemplate is a general text generator for creating topology files for simulation programs (like LAMMPS and ESPResSo). Users can create a molecule "template" (a text file) containing all of the text relevant to a particular molecule. Then moltemplate can duplicate it, customize it, and use it as a building-block for constructing larger molecules. (Moltemplate maintains atom-counts, bond-counts, and any other counters.) Files created by moltemplate can be combined with the coordinates generated by PACKMOL.
An example of a quite sophisticated illustrative molecular structure of a coarse grained spherical lipid bilayer with proteins, built with Packmol, and the corresponding Moltemplate files, was provided by the Moltemplate developer Andrew Jewett:
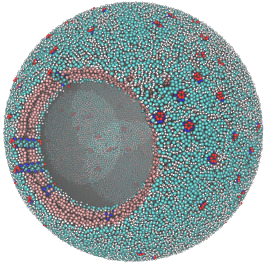 |
Packmol setup files Moltemplate files LAMMPS files |
Avogadro's packmol plugin
Tim Vandermeersch is developing a beautiful Packmol plugin for Avogadro, consisting of a user interface that will help many people to use Packmol. This looks very promising, and we are following closely its development! More at Tim's blog and at the project's source repository.
MolecV1
Iteratively asks properties of the system, as the density and type of box to be created, and writes a input file for packmol. [Click here]