Basic user guide
Here the usage of the functions that allocate the list of distances will be described. Different running modes are available depending on the expected output.
Installation and loading
To install MolecularMinimumDistances, first download and install Julia (1.6 or greater) from https://julialang.org/downloads/. Install and run it. Then, use:
julia> import Pkg; Pkg.add("MolecularMinimumDistances")
julia> using MolecularMinimumDistancesExample input files
The examples here use a molecular system, but the package actually only considers the coordinates of the atoms and the number of atoms of each molecule. Thus, more general distance problems can be tackled.
The input atomic positions used in the following examples can be obtained with:
julia> using MolecularMinimumDistances, PDBTools
julia> system = MolecularMinimumDistances.download_example()
Array{Atoms,1} with 62026 atoms with fields:
index name resname chain resnum residue x y z beta occup model segname index_pdb
1 N ALA A 1 1 -9.229 -14.861 -5.481 0.00 1.00 1 PROT 1
2 HT1 ALA A 1 1 -10.048 -15.427 -5.569 0.00 0.00 1 PROT 2
3 HT2 ALA A 1 1 -9.488 -13.913 -5.295 0.00 0.00 1 PROT 3
⋮
62024 OH2 TIP3 C 9339 19638 13.485 -4.534 -34.438 0.00 1.00 1 WAT2 62024
62025 H1 TIP3 C 9339 19638 13.218 -3.647 -34.453 0.00 1.00 1 WAT2 62025
62026 H2 TIP3 C 9339 19638 12.618 -4.977 -34.303 0.00 1.00 1 WAT2 62026
The system consists of a protein (with 1463 atoms), solvated by 181 TMAO molecules (with 14 atoms each), 19338 water molecules, and some ions.
These coordinates belong to a snapshot of a simulation which was performed with cubic periodic boundary conditions, with a box side of 84.48 Angstrom.
The coordinates of each of the types of molecules can be extracted from the system array of atoms with (using PDBTools - v0.13 or greater):
julia> protein = coor(system,"protein")
1463-element Vector{StaticArrays.SVector{3, Float64}}:
[-9.229, -14.861, -5.481]
[-10.048, -15.427, -5.569]
[-9.488, -13.913, -5.295]
⋮
[6.408, -12.034, -8.343]
[6.017, -10.967, -9.713]
julia> tmao = coor(system,"resname TMAO")
2534-element Vector{StaticArrays.SVector{3, Float64}}:
[-23.532, -9.347, 19.545]
[-23.567, -7.907, 19.381]
[-22.498, -9.702, 20.497]
⋮
[13.564, -16.517, 12.419]
[12.4, -17.811, 12.052]
julia> water = coor(system,"water")
58014-element Vector{StaticArrays.SVector{3, Float64}}:
[-28.223, 19.92, -27.748]
[-27.453, 20.358, -27.476]
[-27.834, 19.111, -28.148]
⋮
[13.218, -3.647, -34.453]
[12.618, -4.977, -34.303]Using these vectors of coordinates, we will illustrate the use of the current package.
Shortest distances from a solute
The simplest usage consists of finding for each molecule of one set the atoms of the other set which are closer to them. For example, here we want the atoms of the proteins which are closer to each TMAO molecule (14 atoms), within a cutoff of 12.0 Angstroms.
The simulations was performed with periodic boundary conditions, in a cubic box of sides [84.48, 84.48, 84.48]. We compute the minimum distances with:
julia> list = minimum_distances(
xpositions=tmao, # solvent
ypositions=protein, # solute
xn_atoms_per_molecule=14,
cutoff=12.0,
unitcell=[84.48, 84.48, 84.48]
)
181-element Vector{MinimumDistance{Float64}}:
MinimumDistance{Float64}(false, 0, 0, Inf)
MinimumDistance{Float64}(false, 0, 0, Inf)
MinimumDistance{Float64}(false, 0, 0, Inf)
⋮
MinimumDistance{Float64}(false, 0, 0, Inf)
MinimumDistance{Float64}(true, 2526, 97, 9.652277658666891)The list contains, for each molecule of TMAO, a MinimumDistance object, containing the following fields, exemplified by printing the last entry of the list:
julia> list[end]
MinimumDistance{Float64}(true, 2526, 97, 9.652277658666891)
Distance within cutoff, within_cutoff = true
x atom of pair, i = 2526
y atom of pair, j = 97
Distance found, d = 9.652277658666891The fields within_cutoff, i, j, and d show if a distance was found within the cutoff, the indexes of the atoms involved in the contact, and their distance.
If the solute has more than one molecule, this will not be taken into consideration in this mode. All molecules will be considered as part of the same structure (the number of atoms per molecule of the protein is not a parameter here).
All shortest distances
A similar call of the previous section can be used to compute, for each molecule of a set of molecules, which is the closest atom of every other molecule of another set.
In the example, we can compute for each TMAO molecule, which is the closest atom of water, and vice-versa. The difference from the previous call is that now wee need to provide the number of atoms of both TMAO and water:
julia> water_list, tmao_list = minimum_distances(
xpositions=water,
ypositions=tmao,
xn_atoms_per_molecule=3,
yn_atoms_per_molecule=14,
unitcell=[84.48, 84.48, 84.48],
cutoff=12.0
);
julia> water_list
19338-element Vector{MinimumDistance{Float64}}:
MinimumDistance{Float64}(true, 2, 1512, 4.779476331147592)
MinimumDistance{Float64}(true, 6, 734, 2.9413928673334357)
MinimumDistance{Float64}(true, 8, 859, 5.701548824661595)
⋮
MinimumDistance{Float64}(true, 58010, 1728, 3.942870781549911)
MinimumDistance{Float64}(true, 58014, 2058, 2.2003220218867936)
julia> tmao_list
181-element Vector{MinimumDistance{Float64}}:
MinimumDistance{Float64}(true, 12, 22520, 2.1985345118965056)
MinimumDistance{Float64}(true, 20, 33586, 2.1942841657360606)
MinimumDistance{Float64}(true, 37, 26415, 2.1992319113726926)
⋮
MinimumDistance{Float64}(true, 2512, 37323, 2.198738501959709)
MinimumDistance{Float64}(true, 2527, 33664, 2.1985044916943015)Two lists were returned, the first containing, for each water molecule, MinimumDistance data associated to the closest TMAO molecule (meaning the atoms involved in the contact and their distance). Similarly, the second list contains, for each TMAO molecule, the MinimumDistance data associated to each TMAO molecule.
Shortest distances within molecules
There is an interface to compute the shortest distances of molecules within a set of molecules. That is, given one group of molecules, compute for each molecule which is the shortest distance among the other molecules of the same type.
A typical call would be:
julia> water_list = minimum_distances(
xpositions=water,
xn_atoms_per_molecule=3,
unitcell=[84.48, 84.48, 84.48],
cutoff=12.0
)
19338-element Vector{MinimumDistance{Float64}}:
MinimumDistance{Float64}(true, 2, 33977, 2.1997806708851724)
MinimumDistance{Float64}(true, 4, 43684, 2.1994928961012814)
MinimumDistance{Float64}(true, 9, 28030, 2.1997583958244142)
⋮
MinimumDistance{Float64}(true, 58010, 22235, 2.1992096307537414)
MinimumDistance{Float64}(true, 58012, 9318, 2.20003227249056)Which contains for each water molecule the atoms involved in the closest contact to any other water molecule, and the distances (within the cutoff). A pictorial representation of a result of this type is, for a simpler system:
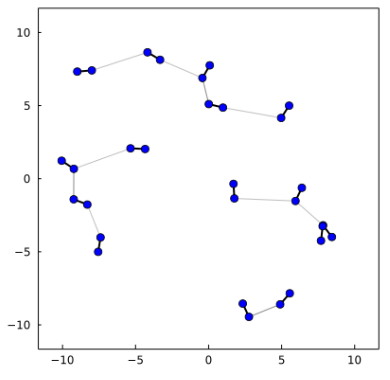
This can be used for the identification of connectivity networks, for example, or for some types of clustering.